Two 2023 papers demonstrated the weak-sauce treatments currently offered to resolve elbow ulnar nerve pain:
“This case report investigated the use of ultrasound-guided nerve hydrodissection and platelet releasate injection for treating ulnar neuritis at the elbow.
- The patient’s symptoms were first managed with home exercise and ulnar nerve hydrodissection at the elbow, which decreased but did not resolve her pain.
- Platelet releasate injection of the ulnar nerve at the elbow was subsequently performed. Six weeks post-procedure, the patient reported additional pain improvement.
Despite these results, the patient was not completely symptom-free. Persistent symptoms were attributed to her concomitant neurogenic thoracic outlet syndrome.”
https://www.cureus.com/articles/133241-platelet-releasate-injection-as-a-novel-treatment-for-ulnar-neuritis-at-the-elbow-a-case-report/ “Platelet Releasate Injection as a Novel Treatment for Ulnar Neuritis at the Elbow: A Case Report”
When a diagnosis concludes with the word ‘syndrome’, we can be assured that medical professionals don’t know any specific cause. Expect physical therapy and/or drugs and/or surgery to be recommended, which will only address symptoms, not causes.
These practitioners proposed two experimental treatments, and somehow, the patient agreed to be a lab rat for both. If they were repeatedly questioned as to whether those two treatments would address causes, I’d expect responses similar to “That’s all we can do for you.”
In line with this decade’s revelations about the medical profession, the patient was also gaslighted. These practitioners asserted “changes to the patient’s lifestyle” as a reason neither treatment worked, although no such lifestyle changes were indicated.
Medical professionals are people whose early life experiences impel them to control other people with a license, among other driving factors. They won’t discuss items outside their ideas and beliefs, because these are defenses against their and their patients’ realities.
Next is a study of 111 elbow neuropathy patients (average age 55, median follow-up period of 880 days), one third of whom had various surgeries:
“There are three main potential mechanisms of recovery after nerve lesion: (1) resolution of conduction block, (2) collateral reinnervation, and (3) nerve regeneration.
- Nerve function in chronic focal compression/entrapment neuropathies seems to improve mainly due to resolution of the conduction block and collateral reinnervation.
- Contribution of nerve regeneration seems to be minor.
The majority of axons lost in chronic focal neuropathies probably never recover. Further studies using quantitative methods are needed to validate present findings.”
https://www.mdpi.com/2077-0383/12/12/3906 “No Major Nerve Regeneration Seems to Occur during Recovery of Ulnar Neuropathy at the Elbow”
Another interesting thing may have unexpectedly started with my 90-day trial of Prodrome Glia and Neuro products. Here’s an abbreviated look at what I’m tracking that omits intermittent fasting data:
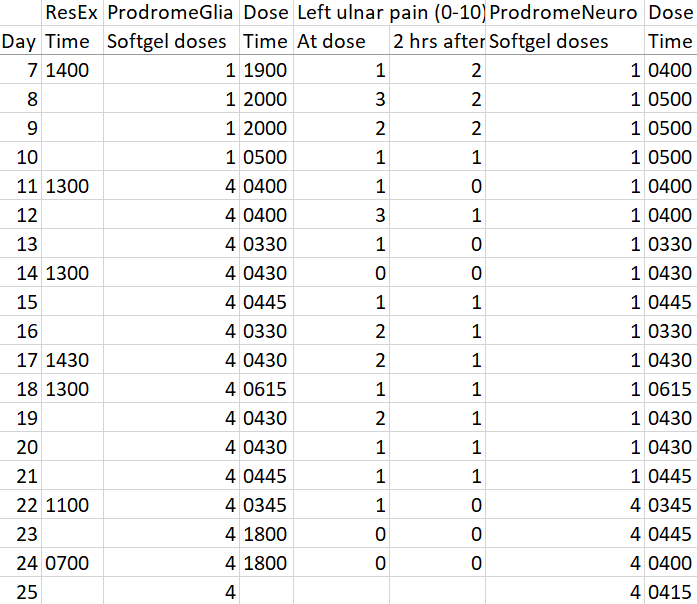
Left ulnar nerve pain stayed the same or decreased two hours after a ProdromeGlia loading dose from Days 11-21. After adding a ProdromeNeuro loading dose at Day 22, my left ulnar nerve pain has unexpectedly stopped.
Any resistance exercise I’ve done during the past month would have aggravated my left ulnar nerve prior to the current regimen. Yesterday I clumped together reverse curls, regular bicep curls, bench presses, and triceps extensions, in that order, two sets each. I used lower weights than in the past, squeezed at the top of concentric motion, and returned slowly with eccentric motion for each rep.
Today on Day 25 the exercised muscles burn as expected, especially due to eccentric motion. But my left ulnar nerve is fine.
At the beginning, I thought that ProdromeGlia might eventually have an effect on left ulnar nerve pain, but not ProdromeNeuro. The first paper noted “The ulnar nerve begins in the axilla as a continuation of the medial cord of the brachial plexus, originally arising from the C8 and T1 nerve roots of the spinal cord.” I’ll guess that something upstream of my left ulnar nerve may also be involved in recent results.
Don’t agree with the second paper’s unevidenced assertion that “The majority of axons lost in chronic focal neuropathies probably never recover.” I’ve had intermittent left ulnar nerve numbness and pain for over five years, which is a lot longer than the 880-day median follow-up period of that paper.
Dr. Goodenowe presented his combined daily plasmalogen precursor dose as ~100 mg/kg. My analogous combined daily plasmalogen precursor loading doses are 7200 mg, appropriate for a person who weighs 72 kg. I weigh 155 lbs. / 70 kg.
More testing is warranted, of course. Maybe I’m just in-between an intermittent occurrence of left ulnar pain. So far, the way my current regimen is playing out, every day has something to make it Thanksgiving Day.




















