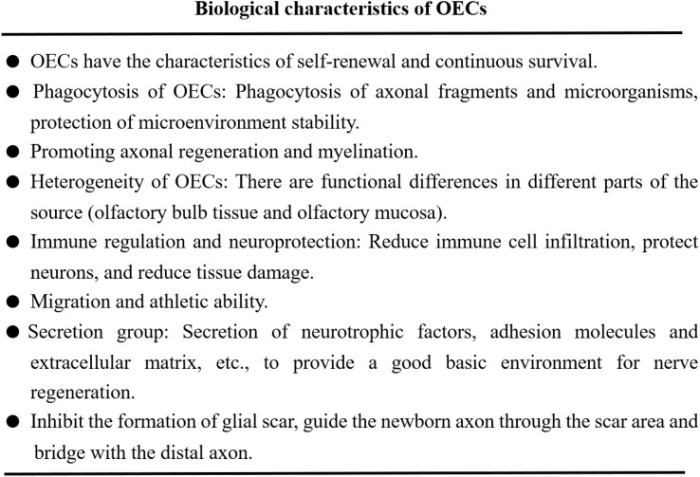
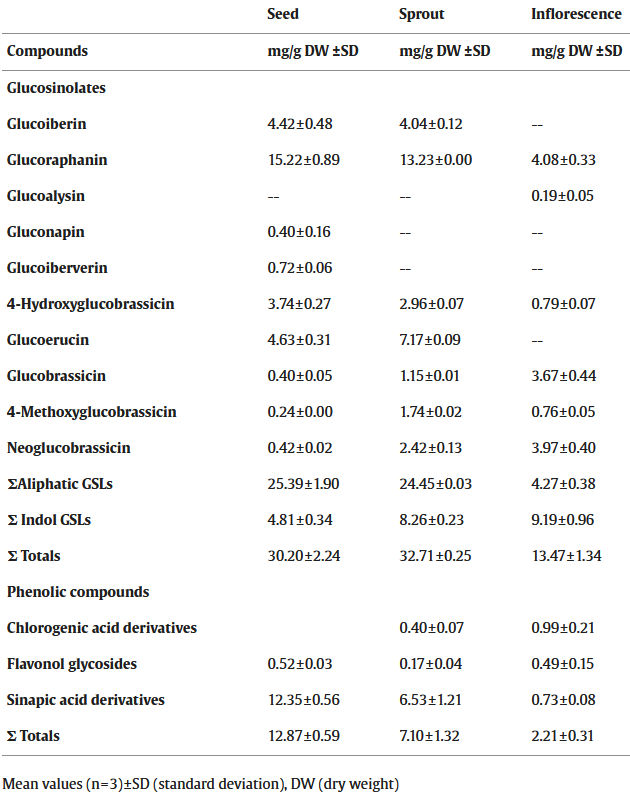
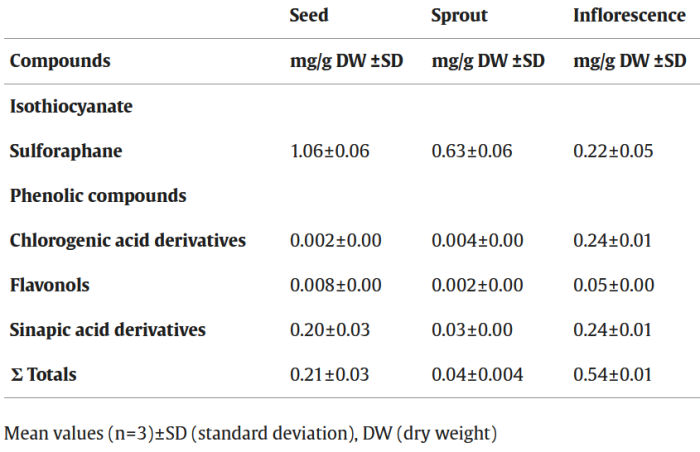
Continuing Part 1 with three 2024 preprint studies, starting with an investigation of neuroinflammation in high school athletes:
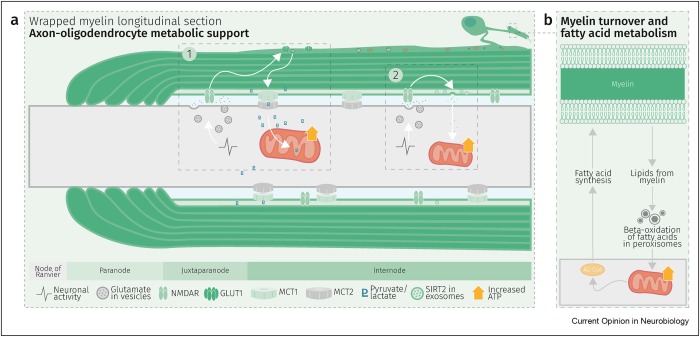
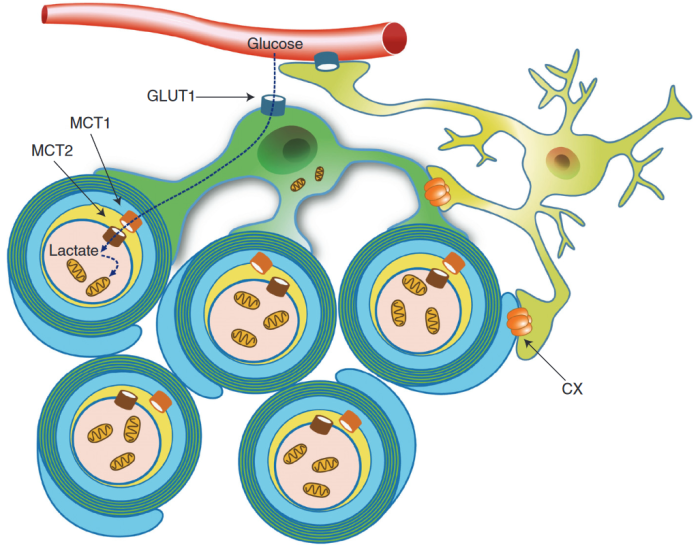
“Axons are long fibers conducting nerve impulses from nerve cells to synaptic ends. Like electric wires, axons are insulated by the myelin sheath produced by oligodendrocytes (ODC) in the brain or Schwann cells in the periphery. The myelin sheath is vulnerable to mechanical stresses after head injuries, as well as targets for autoimmune attack in multiple sclerosis and degeneration in various white matter diseases.
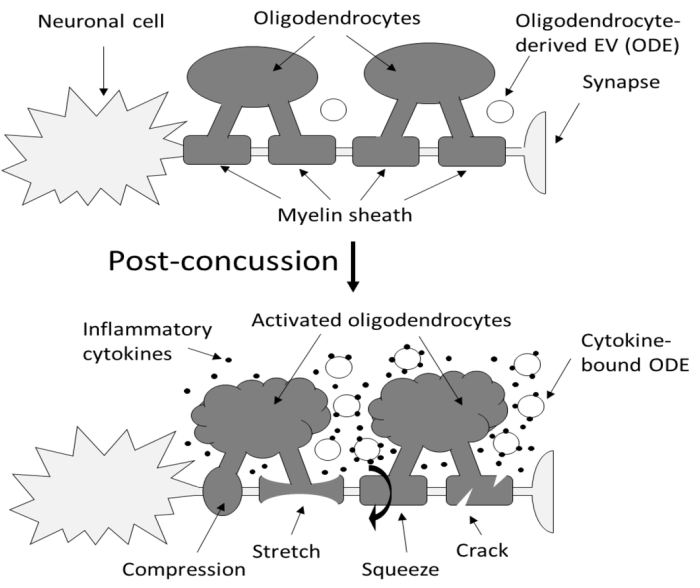
It is challenging to definitively validate axonal neuroinflammation, because axonal neuroinflammation is only diagnosed at post-mortem autopsy, or wait for more than a decade to potentially witness progression to chronic traumatic encephalopathy, or white matter dementia. Advanced imaging analysis of computed tomography and magnetic resonance imaging are not sensitive enough to identify such microscopic abnormalities.
We developed a sandwich immunoassay detecting dual signals of myelin oligodendrocyte glycoprotein (MOG) and interleukin 1B (IL1B) in human plasma, [IL1B on MOG]. MOG is a transmembrane protein specifically expressed in ODC and Schwann cells membranes, and doesn’t freely exist in plasma. We found that serum from capillary blood is acceptable, and we tested control and athlete samples using only 5 mL samples. When we tested 63 control plasma samples, values were widely distributed over 2 logs, so we focused on longitudinal studies.
Damaged neurons are not easily detectable using conventional physical examinations, because the brain’s inherent adaptability allows it to compensate for localized damage by finding alternate routes. While this adaptability is advantageous, it also means that these concealed lesions can go unnoticed, potentially leading to future complications.
Elevation of [IL1B on MOG] was seen in some athletes who did not show concussion or traumatic brain injury (TBI). While the occurrence of concussion is relatively limited, potential prevalence of subconcussion or subconcussive condition is expected to be substantially higher.
If [IL1B on MOG] levels remain unchanged during this early post-concussion period (2-4 weeks), it may suggest that neuroinflammation has not been induced, potentially providing reassurance for the athletes to return to play. Conversely, if [IL1B on MOG] levels increase within this timeframe, it may indicate the need for intervention or closer monitoring. Thus, there is compelling potential for incorporating this test into concussion guidelines.”
https://www.researchsquare.com/article/rs-3997676/v1 “An approach for the analysis of axonal neuroinflammation by measuring dual biomarkers of oligodendrocytes and inflammatory cytokine in human plasma”
A rodent study investigated the immune system’s influence on oligodendrocyte lineage cells after TBI:
“White matter injury is thought to be a major contributor to long-term cognitive dysfunctions after TBI. This damage occurs partly due to apoptotic death of oligodendrocyte lineage cells (OLCs) after injury, triggered directly by the trauma or in response to degenerating axons.
Our data indicates that depletion of the gut microbiota after TBI impaired remyelination, reduced OLCs proliferation, and required the presence of T cells. This suggests that T cells are an important mechanistic link by which the gut microbiota modulate oligodendrocyte response and white matter recovery after TBI.
Our findings suggest that oligodendrocytes are not passive in the neuroinflammatory and degenerative environment caused by brain trauma, but instead could exert an active role in modulation of immune response.”
https://www.researchsquare.com/article/rs-4289147/v1 “Gut Microbiota Shape Oligodendrocyte Response after Traumatic Brain Injury”
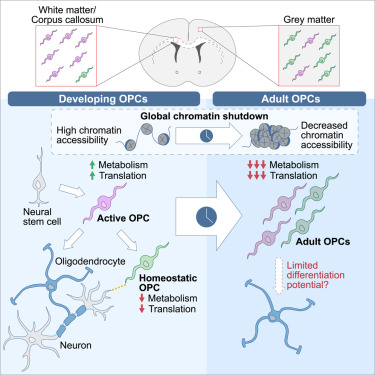
A rodent study investigated whether oligodendrocyte precursor cells had myelination-independent roles in brain aging:
“OPCs, the source cells of myelin-forming cells in the central nervous system, have been linked to brain aging by their compromised differentiation and regeneration capability. Our results demonstrate that macroautophagy influx declines in aged OPCs, which results in the accumulation of senescent OPCs in aged brains. Senescent OPCs impair neuronal plasticity and exacerbate neurodegeneration, eventually leading to cognitive decline.
Inactivation of autophagy in OPCs exhibits a limited effect on myelin thickness but a loss of myelin in middle-aged mice. The loss of myelin observed is an adaptational change to suppressed neuronal plasticity. However, neither the number of OLs nor oligodendrogenesis is altered by inactivation of autophagy in adult OPCs.
The present study indicates that the intervention of senescent OPCs is an additional promising therapeutic strategy for aging and aging-related cognitive deficits. Autophagy regulates senescence by impairing protein turnover, mitochondrial homeostasis, oxidative stress, and maintaining senescence-associated secretory phenotype. Further investigation remains on whether autophagy in OPCs shares the exact mechanism to promote senescence as that in other types of cells.
Considering autophagy declines with aging, our study brings a novel mechanism in brain aging. Declined autophagy causes senescence of OPCs, which impairs neuronal plasticity and exacerbates neurodegeneration via CCL3/5-CCR5 signaling.”
https://www.researchsquare.com/article/rs-3926942/v1 “Impaired Macroautophagy in Oligodendrocyte Precursor Cells Exacerbates Aging-related Cognitive Deficits via a Senescence Associated Signaling”