In this 2023 presentation for a professional audience, Dr. Dayan Goodenowe showed an example of what could be done (in the form of what he personally did at ages 53-54) to restore and augment brain structure and function over a 17-month period by taking plasmalogens and supporting supplements:
Follow the video along with its interactive transcript. Restorative / augmentative supplements included:

Forms of MRI used to document brain structure and function changes were:

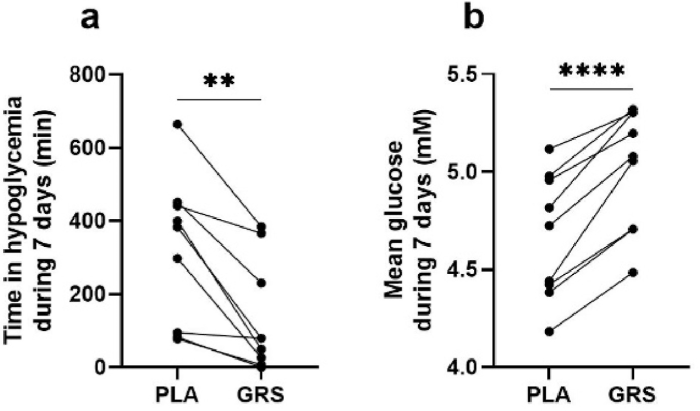
Brain volume decreases are the rule for humans beginning at age 40. Dr. Goodenowe documented brain volume increases, which aren’t supposed to happen, but did per the below slide of overall results:

“From a global cortical volume and thickness perspective, 17 months of high-dose plasmalogens reversed ~15 years of predicted brain deterioration.”
Specific increased adaptations in brain measurements over 17 months included:
- Cortical thickness .07/2.51 = +3%.
- White matter microstructure fractional anisotropy +8%.
- Nucleus accumbens volume +30%.
- Dopaminergic striatal terminal fields’ volume +18%.
- Cholinergic cortical terminal fields’ volume +10%.
- Occipital cortex volume +10%.
- Optic chiasm volume +225%.
- Nucleus basalis connectivity.
- Neurovascular coupling signal controlled by noradrenaline integrity.
- Amygdala volume +4% and its connectivity to the insula, indicating ongoing anxiety and emotional stress response.
- Parahippocampus volume +7%.
- Hippocampus fractional anisotropy +5%.
No changes:
- Amygdala connectivity to the ventral lateral prefrontal cortex, the same part of the brain that relates to placebo effect.
- Hippocampus connectivity.
Decreased adaptations in brain measurements included:
- White matter microstructure radial diffusivity -10%.
- Amygdala connectivity to the anterior cingulate cortex to suppress / ignore / deny anxiety response.
- Amygdala connectivity to the dorsal lateral prefrontal cortex.
- Entorhinal cortex volume -14%.
- Hippocampus volume -6%.
- Hippocampus mean diffusivity (white matter improved, with more and tighter myelin) -4%.
The other half of this video was a lively and wide-ranging Q&A session.
The referenced 2023 study of 653 adults followed over ten years showed what brain deterioration could be expected with no interventions. Consider these annual volume decrease rates to be a sample of a control group:

https://jamanetwork.com/journals/jamanetworkopen/fullarticle/2806488 “Characterization of Brain Volume Changes in Aging Individuals With Normal Cognition Using Serial Magnetic Resonance Imaging”
Also see a different population’s brain shrinkage data in Prevent your brain from shrinking.
The daily plasmalogen precursor doses Dr. Goodenowe took were equivalent to 100 mg softgel/kg, double the maximum dose of 50 mg softgel/kg provided during the 2022 clinical trial of cognitively impaired old people referenced in Plasmalogens Parts 1, 2, and 3.
He mentions taking 5 ml in the morning and 5 ml at night because he used the Prodrome oil products. 1 ml of a Prodrome oil plasmalogen precursor product equals 900 mg of their softgel product.
“My brain is trying to minimize long-term effects of pain/stress by suppressing my memory of it. But this can only go on for so long before it becomes an entrenched state.
I have solved the sustenance side of the equation. I need to work harder to solve the environmental side.”
While I agree that we each have a responsibility to ourselves to create an environment that’s conducive to our health, the above phenomenon isn’t necessarily resolvable by changing an individual’s current environment. My understanding is that long-term effects of pain, stress, and related human experiences are usually symptoms of causes that started much earlier in our lives.
Adjusting one’s present environment may have immediate results, but probably won’t have much therapeutic impact on long-term issues. Early life memories and experiences are where we have to gradually go in order to stop being driven by what happened back then.
See Dr. Arthur Janov’s Primal Therapy for its principles and explanations. I started Primal Therapy at a similar age, 53, and continued for three years.