This 2021 computational study investigated several methods of improving epigenetic clock reliability:
“Epigenetic clocks are widely used aging biomarkers calculated from DNA methylation data. Unfortunately, measurements for individual CpGs can be surprisingly unreliable due to technical noise, and this may limit the utility of epigenetic clocks.
Noise produces deviations up to 3 to 9 years between technical replicates for six major epigenetic clocks. Elimination of low-reliability CpGs does not ameliorate this issue.
We present a novel computational multi-step solution to address this noise, involving performing principal component analysis (PCA) on the CpG-level data followed by biological age prediction using principal components as input. This method extracts shared systematic variation in DNAm while minimizing random noise from individual CpGs.
Our novel principal-component versions of six clocks show agreement between most technical replicates within 0 to 1.5 years, equivalent or improved prediction of outcomes, and more stable trajectories in longitudinal studies and cell culture. This method entails only one additional step compared to traditional clocks, does not require prior knowledge of CpG reliabilities, and can improve the reliability of any existing or future epigenetic biomarker.
PC-based clocks showed greatly improved agreement between technical replicates, with 90+% agreeing within 1-1.5 years. The median deviation ranged from 0.3 to 0.8 years, whereas CpG clocks ranged from 0.9-2.4 years.
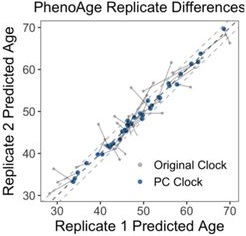
The most dramatic improvement was in PhenoAge. CpG-trained PhenoAge has a median deviation of 2.4 years, 3rd quartile of 5 years, and maximum of 8.6 years. In contrast, PCPhenoAge has a median deviation of 0.6 years, 3rd quartile of 0.9 years, and maximum of 1.6 years. PCPhenoAge was trained directly on phenotypic age based on clinical biomarkers rather than DNAm.
Correlations between different PC clocks was stronger than between CpG clocks. This may be partly due to the shared set of CpGs used to train PCs, or due to the reduction of noise that would have biased correlations towards the null. Correlations between PC clocks and CpG clocks tended to be stronger compared to correlations between CpG clocks and CpG clocks, consistent with a reduction of noise.
PC clocks preserve relevant aging signals unique to each of their CpG counterparts. They reduce technical variance but maintain relevant biological variance.
PCA is a commonly used tool and does not require specialized knowledge. High reliability of principal component-based epigenetic clocks will make them particularly useful for applications in personalized medicine and clinical trials evaluating novel aging interventions.”
https://doi.org/10.1093/geroni/igab046.015 “A Computational Solution to Bolster Epigenetic Clock Reliability for Clinical Trials and Longitudinal Tracking”
I appreciate that a coauthor – who is the originator of PhenoAge – is open to evidence and improvements. There’s a fun do-it-yourself demo of PCA at https://setosa.io/ev/principal-component-analysis/.
I found this study from it citing a 2021 review:
https://www.sciencedirect.com/science/article/abs/pii/S1084952121000094 “Aging biomarkers and the brain” (not freely available)
I found that review from it citing a 2020 study:
https://www.cell.com/iscience/fulltext/S2589-0042(20)30384-9 “Human Gut Microbiome Aging Clock Based on Taxonomic Profiling and Deep Learning”
Maybe this last study could be improved from its “mean absolute error of 5.91 years” with PCA? See Part 2 for another view.