Here are two 2025 papers, starting with a rodent study that investigated interactions between the Nrf2 and kynurenine pathways:
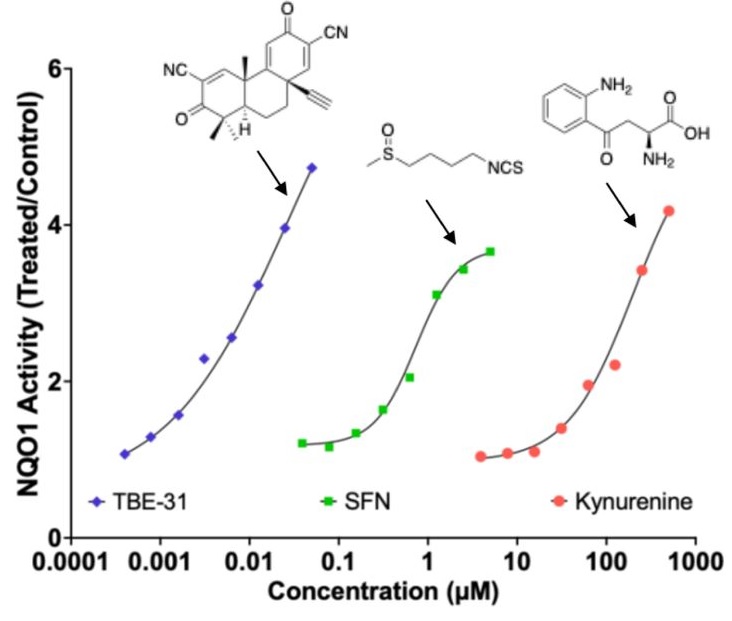
“Exposure to the tryptophan metabolite kynurenine and its electrophilic derivative kynurenine-carboxyketoalkene (Kyn-CKA) leads to an increase in the abundance of transcription factor Nrf2 and induction of Nrf2-target genes. The Keap1/Nrf2 system is the main orchestrator of cellular defence against environmental stress, most notably oxidative and inflammatory stress.
Nrf2 can be activated pharmacologically by small molecules, the majority of which are electrophiles and oxidants that modify specific cysteine-based sensors in Keap1. C151 in Keap1 is the target of the isothiocyanate sulforaphane, a classical Nrf2 activator that has been employed in ∼90 clinical trials, as well as for the two Nrf2 activators that are clinically in use: dimethyl fumarate, for relapsing remitting multiple sclerosis, and omaveloxolone, for Friedreich’s ataxia.
Kynurenine is an endogenous metabolite derived from the essential amino acid tryptophan. Kynurenine and its metabolites, such as the electrophilic kynurenine-carboxyketoalkene (Kyn-CKA), have been demonstrated to activate Nrf2 in other pathologies, including sickle cell disease, attenuating inflammation. Moreover, identification of the gene encoding the kynurenine-metabolising enzyme kynureninase as a gene transcriptionally upregulated by Nrf2, provides a plausible negative feedback regulatory mechanism.
Because kynurenine is not electrophilic, whereas its metabolite Kyn-CKA is, we considered the possibility that Kyn-CKA is the actual Nrf2 activator. Using biochemical and cell-based assays, we found that Kyn-CKA reacts with C151 in the BTB domain of Keap1 and increases the thermostability of Keap1, indicating target engagement. Consequently, Nrf2 accumulates and induces transcription of antioxidant/electrophile-responsive element (ARE/EpRE)-driven genes.
These findings demonstrate that Kyn-CKA targets C151 in Keap1 to derepress Nrf2, and reveal that Nrf2 is a main contributor to the anti-inflammatory activity of Kyn-CKA in macrophages.”
https://www.sciencedirect.com/science/article/pii/S2213231726000078 “The electrophilic metabolite of kynurenine, kynurenine-CKA, targets C151 in Keap1 to derepress Nrf2”
A review subject was targeting nicotinamide adenine dinucleotide, oxidized form (NAD+) for clinical use:
“Mammalian NAD+ biosynthesis includes four known pathways, primarily occurring in cytoplasm:
- (a) the NRH pathway;
- (b) the salvage pathway;
- (c) the Preiss–Handler pathway; and
- (d) the kynurenine pathway.
The de novo kynurenine pathway metabolizes tryptophan (Trp) to NAD+, producing various intermediates that serve as biomarkers for different diseases. These intermediates show alterations in various pathological conditions.
While kynurenine and its metabolic derivatives are intermediates in the de novo NAD+ biosynthesis pathway, these are also produced independently in various physiological contexts, particularly in immune cells, where they act as immunomodulatory compounds.”
https://www.nature.com/articles/s43587-025-00947-6 “Emerging strategies, applications and challenges of targeting NAD+ in the clinic” (not freely available) Thanks to Dr. Jianying Zhang for providing a copy.
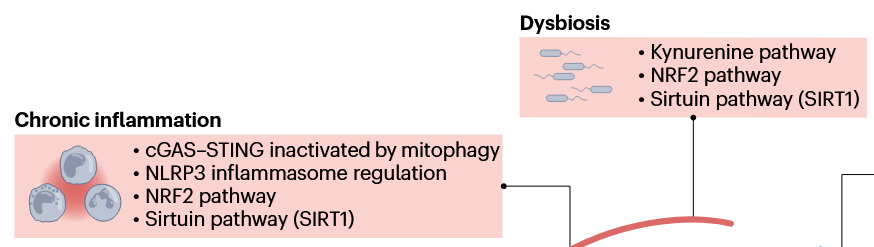
This second paper above showed a graphic of the Nrf2 and kynurenine pathways together in a diagram showing relationships between NAD+ augmentation and the hallmarks of aging, but didn’t elaborate other than labeling their box Dysbiosis. So how these two pathways interact is better outlined in the first paper above with explaining how a kynurenine-metabolizing enzyme is one of the hundreds of Nrf2 target genes, creating a natural feedback loop between Nrf2 activation and the kynurenine pathway.
These reviewers also lumped SIRT1 in their Dysbiosis box, and into several other boxes, probably due to the penultimate coauthor’s influence:
However, repeating something over and over doesn’t make it scientifically valid regardless of the number of citations. Or, as a 2022 review Sirtuins are not conserved longevity genes concluded:
“A global pursuit of longevity phenotypes was driven by a mixture of framing bias, confirmation bias, and hype. Review articles that propagate these biases are so rampant that few investigators have considered how weak the case ever was for sirtuins as longevity genes.
Acknowledging that a few positive associations between sirtuins and longevity have been identified after thousands of person-years and billions of dollars of effort, we review the data and suggest rejection of the notions that sirtuins (i) have any specific connection to lifespan in animals and (ii) are primary mediators of the beneficial effects of NAD repletion.”